Preparative electrophoresis systems for targeted size selection of biomolecules.
Accelerate to discover
Related topics
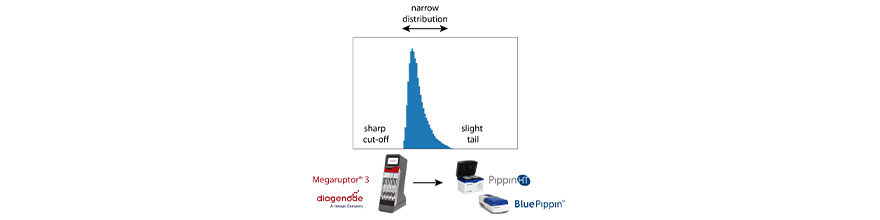
“Range+T “ for Tight Sizing of HMW Libraries
Sage’s DNA size selection technology has been used to help improve PacBio sequencing, particularly for mammalian and complex genome work since the introduction of the BluePippin (2012). Over the last few years, PacBio®’s Hi-Fi circular consensus sequencing has been the method that has set the gold standard for whole genome sequencing. In terms of library size, this technique benefits greatly from a sharp size cut-off at the LMW end of the size distribution (~10kb) and a slight tail at the HMW end. It also benefits from a narrow fragment size distribution around 17-18kb. A common way to accomplish this is to shear DNA with Diagenode’s Megaruptor®3 device, followed by size selection on a PippinHT or BluePippin.
Range+T is available now for both PippinHT and BluePippin. It requires a software upgrade (v.6.41 and v1.14, respectively, available for download on sagescience/support). We have also packaged cassette kits specifically for this purpose; HRT7510 and BRT7510. Starting Range values can be between 9-30kb.
Related technologies: “Range+T “ for Tight Sizing of HMW Libraries